This content originally appeared on Level Up Coding - Medium and was authored by Ronny Polle
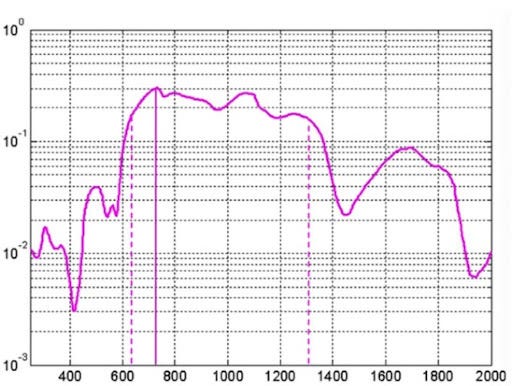
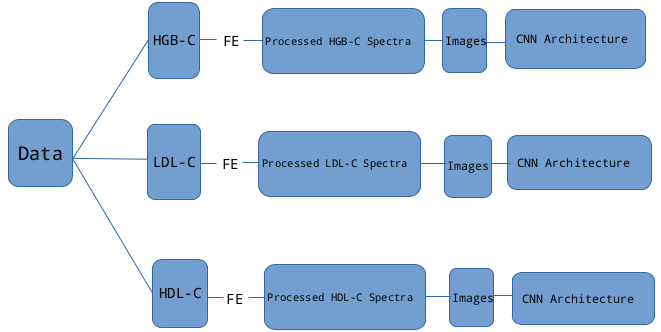
Blood analysis in the clinical settings is done by collecting blood samples from different patients, followed by conducting different blood tests. Using NIR spectroscopy, a beam of light in a given range of wavelengths is directed towards a sample. Depending on the chemical composition (molecular structure) of a sample, light is attenuated at varying degrees — partially absorbed or reflected. Spectroscopic absorption intensity data can be acquired across different wavelengths (biological window) characterizing the chemical composition of biological samples.
Zindi Problem Definition
“For this purpose, you build machine learning models that can classify the level of specific chemical compounds in samples from their spectroscopic data.” ~ Zindi
Data Processing Pipeline
One key challenge in this type of problem is the duplicated readings in addition to high feature dimensionality of spectroscopic readings.
To save compute resources, the median values of absorbance intensities were aggregrated across all sample donation ids. This allowed de-duplication of readings and hence resulting in reduced sample sizes(train + test).
The median aggregation method outperformed mean aggregation technique as demonstrated by experimental results (my hypothesis is that, this might be as a result of the presence of outliers and high variance in the data).
Data Preparation and Spectral Pretreatment
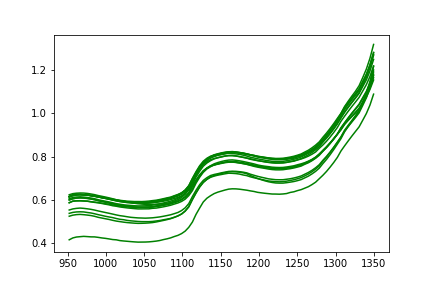
Using the chemical signatures of the pure compounds, signature chemical patterns were extracted from the full readings by computing the correlation of all pure chemical signatures against full spectral readings.
Since hemoglobin is a primary carrier of majority of chemical compounds in the blood , signal intensities observed in compounds was higher compared to the chemical constituents of interest.
Also , a high degree signal overlap is observed at specific wavelengths making the problem of wavelength selection and constituent isolation trickier.
Chemical Constituent Pattern Information
My approach to isolating individual chemical constituents using their corresponding pure chemical signatures involves computing and subtracting correlation signal information of interfering compounds from correlation signal of Hemoglobin( the primary carrier)
from scipy import signal
def compute_correlation(whole, sub):
""" compute correlation matrix
whole : spectral reading
sub : pure chemical signature
"""
whole_mean = whole.mean()
whole = whole - whole_mean
sub = sub - whole_mean
corr = signal.correlate(whole, sub, mode = 'same')
return corr
To obtain an effective metabolite information with a high signal-to-noise ratio, all the pure chemical signature correlation information against the readings were utilized in the computation.
High Density Lipoprotein Cholesterol (HDL-C) signal : To obtain net HDL-C signal intensity information, glucose and fat correlation signals were subtracted from the Hemoglobin correlation matrix per reading.
net_hdl_corr = hgb_corr - (fat_corr + glu_corr)
Low Density Lipoprotein Cholesterol (LDL-C signal ): To obtain the net LDL-C signal intensity information, only glucose correlation is removed from hemoglobin correlation matrix ,per reading. Fat is treated differently in this situation because a high fat content is highly correlated with a high LDL-C. Fat(Triglycerides) can be viewed as an essential precursor for LDL-C synthesis.
net_ldl_corr = ldl_corr - glu_corr
Hemogblobin (HGB) signal : To obtain the net HGB-C signal intensity information, glucose and fat is removed from the global Hemoglobin correlation matrix per reading ( similar for HDL-C )
net_hgb_corr = hgb_corr - (fat_corr + glu_corr)
Experimental spectral pretreatment summary and some key takeaways
- Selecting the “right” sequence of signal pretreatment effects to apply to the data
— Experimental results revealed that performing a sequence of Standard Normal Variate(SNV) scaling followed by Extended Mean Scatter Correction(EMSC) scaling gave better results.
- Applying first order derivative Savitsky-Golay(SG) smoothing to the pretreated signals offered best results.
— Experimental results revealed that the 2nd order SG smoothing introduced too much noise and hence led to worst performance than observed with 1st order SG smoothing
— Basic smoothing effect (without derivatives) and raw specral information offered no improvement.
— Wavelength selection using Partial Least Squares Regression(PLSR) added no improvement
— Finally, data augmentation(before transforming to image data) added no improvement to the model performance.
Converting Spectral Information to Images
- Processed signal information is encoded as different types of images , namely, Grammian Angular Summation Fields(GASF) and Grammian Angular Difference Fields(GADF), and Markov Transition Fields (MTF)
- These imaging techniques involve representing the net signal information in a polar coordinate system rather than the classical cartesian system
- The 3 different images computed from the processed signals are concatenated to form a 3-channel compound image
- For a detailed explanation , consider reading this paper .
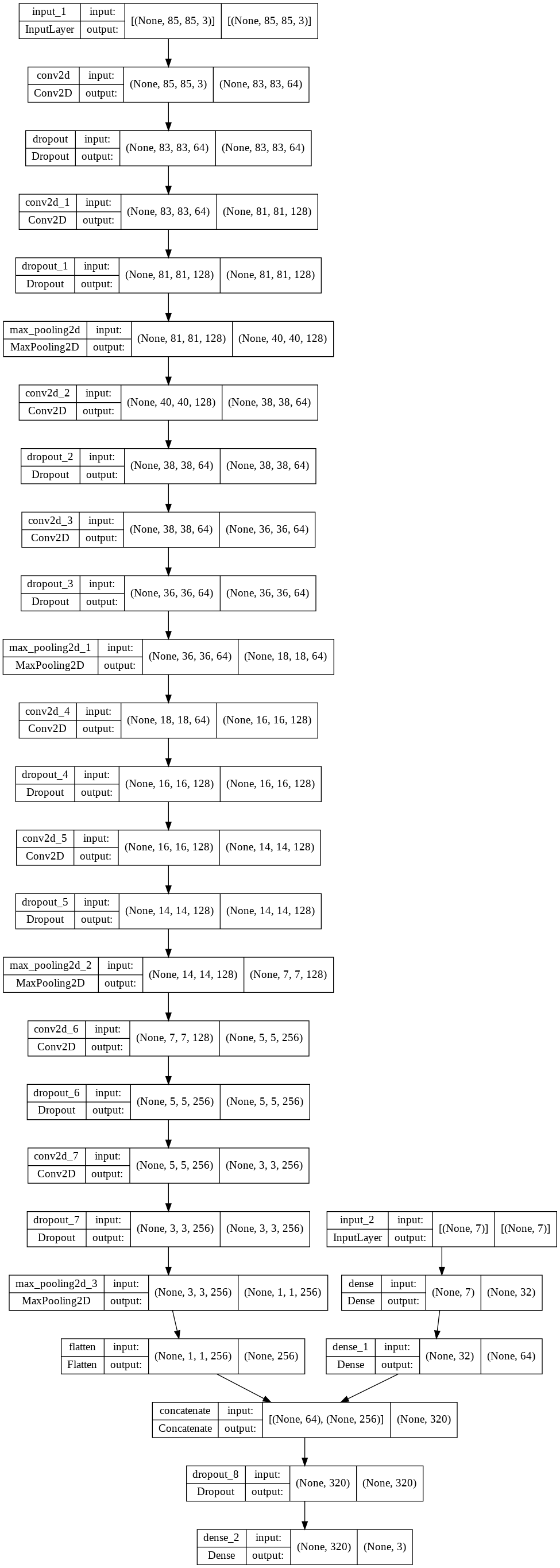
Model Architecture
- Model is composed of a VGG-style model architecture which takes 2 inputs — image and meta features
- Meta features includes the scanner environment variables (temperature + humidity) in addition to engineered features — clusters, principal components, and standard deviation of 1st order SG smoothed spectra.
- Softmax is the activation function utilized across all convolutional hidden layers.
Training
- A uniform model architecture design is maintained during the training phase for the 3 different datasets.
- 10-fold stratified cross validation scheme is utilized per dataset type per model.
In Conclusion
Final Remarks : An appropriate combination of pretreatment of NIR spectroscopic data of blood samples ,and spectral transformation to images can benefit the advantages of image recognition using 2-dimensional Convolutional Neural Networks. This apporach yielded a best accuracy score of 0.6095 - about 8.5% improvement over the benchmark accuracy of 0.5616.
References
[1] My repository — (https://github.com/DrCod/Blood-Spectroscopy-Classification-using-Deep-Learning). Find link to full solution here.
[2] Imaging Time-Series to Improve Classification and Imputation (https://www.ijcai.org/Proceedings/15/Papers/553.pdf) — I highly recommend reading this paper.
[3] pyts — an open source python package for time series classification.
[4] ChemUtils — an open-source python utilities for chemometric data pre-processing and analysis.
[5] Time Series Classification(TSC) Exploration with LSTMs and Convolutional Neural Networks(CNNs) by Rohan (https://www.kaggle.com/ubitquitin/tsc-exploration)
[6] Extended Multiplicative Signal Augmentation(EMSA) — The model implementation structure was adapted from a VGG-style architecture found here
[7] Classification of cartilage integrity based on NIR spectroscopy(https://github.com/ioafara/ML-DL-NIR-spectral-analysis)
[8] bloods.ai Blood Spectroscopy Classification Challenge (https://zindi.africa/competitions/bloodsai-blood-spectroscopy-classification-challenge)
Blood Spectroscopy to Image Classification was originally published in Level Up Coding on Medium, where people are continuing the conversation by highlighting and responding to this story.
This content originally appeared on Level Up Coding - Medium and was authored by Ronny Polle
Ronny Polle | Sciencx (2022-03-19T21:14:41+00:00) Blood Spectroscopy to Image Classification. Retrieved from https://www.scien.cx/2022/03/19/blood-spectroscopy-to-image-classification/
Please log in to upload a file.
There are no updates yet.
Click the Upload button above to add an update.
