This content originally appeared on HackerNoon and was authored by Homology
:::info Authors:
(1) STUART KAUFFMAN;
(2) ANDREA ROL.
:::
Table of Links
Part II. The first Miracle: The emergence of life is an expected phase transition – TAP and RAF.
Part IV. New Observations and Experiments: Is There Life in the Cosmos?
Conclusion and Acknowledgments
FIGURES
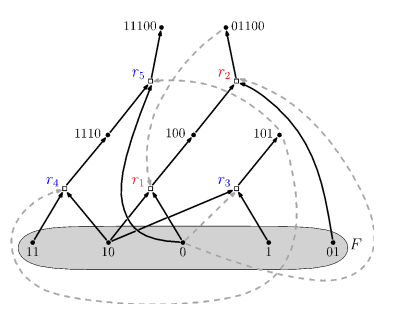
\ A simple collectively autocatalytic set. The model molecules are bit strings acting as substrates and products of reactions. Black solid arrows are drawn from the dots representing substrates of a reaction to a box representing the reaction. Black solid arrows are drawn from the reaction box to the dots representing the products of the reactions. The actual direction of flow of the reaction depends upon displacement from equilibrium. Dashed lines from dots representing molecules to the boxes representing reactions depict which molecules catalyze which reactions.
\ The exogenously supplied food set of monomers and dimers is shown in the grey oval. Derived from (28).
\
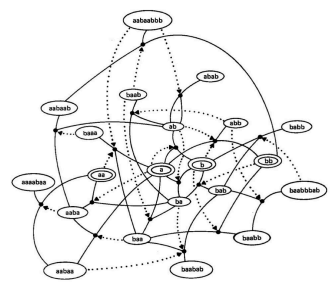
\ A collectively autocatalytic set of linear polymers derived from reference (3). Ovals contain polymers of two monomer types, A and B. Allowed reactions, shown as dots, are cleavage and ligation reactions. A dotted arrow from a molecule oval to a reaction dot indicates that that molecule catalyzes that reaction. Derived from (3).
\
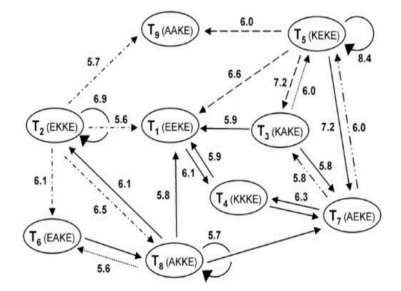
\ The nine peptide collectively autocatalytic set discussed in reference (9). The ovals show the molecules, the arrows show the transitions among the molecules and the relative rates.
\
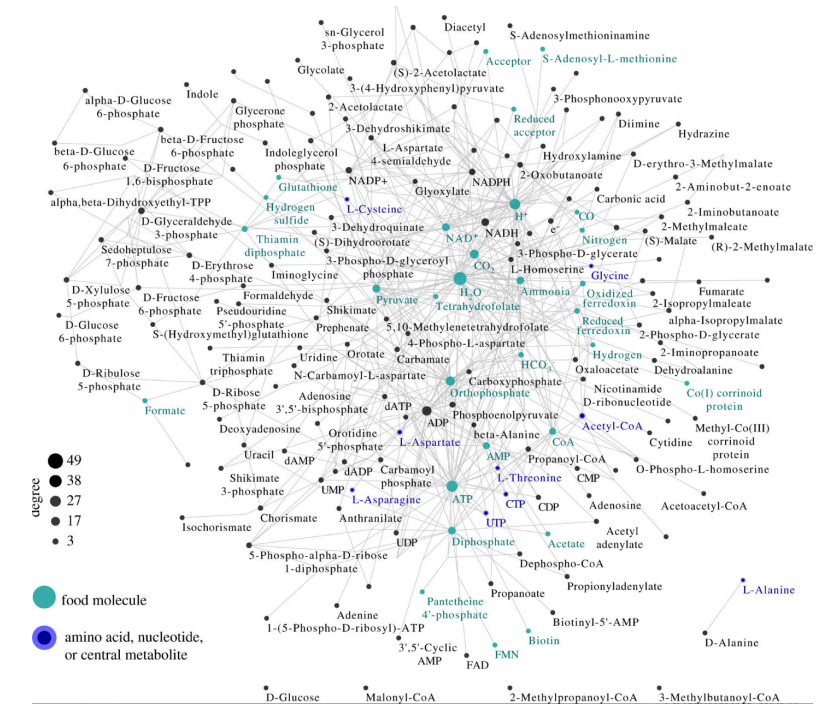
\ A small molecule collectively autocatalytic set with no DNA, RNA, or peptide polymers in a prokaryote. Similar small molecule autocatalytic sets are found in all 6700 prokaryotes. Presumably the phylogeny among these is part of the evolution of metabolism.
\
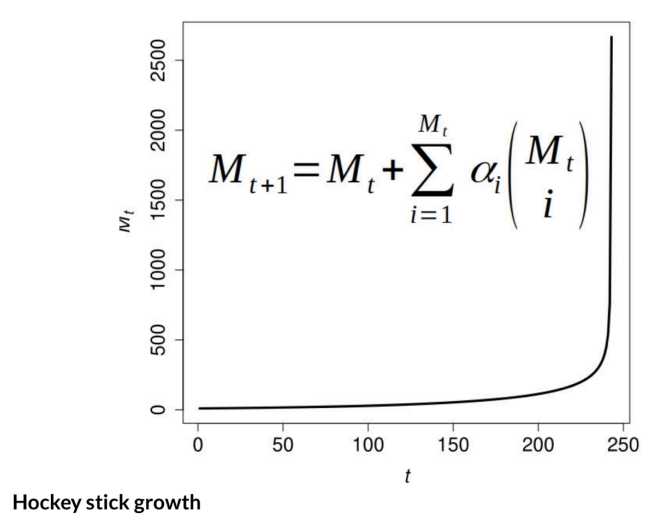
\ The Theory of the Adjacent Possible (TAP) equation and its dynamics. Thanks to W. Hordijk.
References
Schrödinger, E. (1992) What is life?: With mind and matter and autobiographical sketches. Cambridge University Press.
\
Hordijk, W. (2013) Autocatalytic sets: From the origin of life to the economy. BioScience 63, no. 11, 877-881.
\
Farmer, J. D., Kauffman, S. A. and Packard, N. H. (1986) Autocatalytic Replication of Polymers. Physica D 2, 50-67.
\
Orgel, L. E. The origin of life on the earth. (1994) Scientific American 271, no. 4 76-83.
\
Paul, N. and G. F. Joyce. (2004) Minimal self-replicating systems. Current opinion in chemical biology 8, no. 6, 634-639.
\
Szostak, J. W. (2012) The eightfold path to non-enzymatic RNA replication. Journal of Systems Chemistry 3, 1-14.
\
Sievers, D. and von Kiedrowski, G. (1998) Self‐replication of hexadeoxynucleotide analogues: autocatalysis versus cross‐catalysis. Chemistry–A European Journal 4, no. 4, 629-641.
\
Vaidya, N., Manapat, M. L., Chen, I. A., Xulvi-Brunet, R., Hayden, E. J. and Lehman, N. E. (2012) Spontaneous network formation among cooperative RNA replicators. Nature 491, no. 7422, 72-77.
\
Wagner, N. and Ashkenasy, G. (2015) How catalytic order drives the complexification of molecular replication networks. Israel Journal of Chemistry 55, no. 8, 880-890.
\
Lancet, D., Zidovetzki, R. and Markovitch, O. (2018) Systems protobiology: origin of life in lipid catalytic networks. Journal of The Royal Society Interface 15, no. 144, 20180159.
\
Xaiver, J., Hordijk, W., Kauffman, S. A., Steel, M., Martin, W. (2020), Autocatalytic chemical networks at the origin of metabolism Proc Roy Soc B, 287: 20192377. https:/doi.org/10.1098/rspb.2019.2377
\
Xavier, J. and Kauffman, S. A., (2022). Small-molecule autocatalytic networks are universal metabolic fossils. Proc Roy Soc A. 380:20210244
\
Kant, I. (2017) Critique of Judgement by Immanuel Kant-Delphi Classics (Illustrated). Vol. 11. Delphi Classics.
\
Kauffman, S. A. (1971). Cellular Homeostasis, Epigenesis, and Replication in Randomly Aggregated Macromolecular Systems. Journal of Cybernetics 1, 71- 96.
\
Kauffman, S. A. (1986). Autocatalytic Sets of Proteins. J Theor Biol 119, 1-24.
\
Hordijk, W., Hein, J. and Mike Steel. (2010) Autocatalytic sets and the origin of life. Entropy 12, no. 7 1733-1742.
\
Montévil, M. and Mossio, M. (2015) Biological organisation as closure of constraints. Journal of theoretical biology 372 179-191.
\
Atkins, P.W. (1984) The Second Law, Scientific American Library, NY.
\
Kauffman, S. A. (2000) Investigations, Oxford University Press, NY.
\
Von Neumann, J. (1966) Theory of self-reproducing automata. Arthur W. Burks, ed. University of Illinois Press.
\
Davies, P. (2019) The Demon in the machine: How hidden webs of information are solving the mystery of life. University of Chicago Press.
\
Margulis, L. (1971) Symbiosis and evolution. Scientific American225, no. 2, 48-61.
\
Margulis , L. and Sagan, D. (2000) What is Life? University of California Press.
\
Campbell, N. A., Williamson, B., Heyden, R. (2006) Biology: Exploring Life. Boston, Massachusetts: Pearson Prentice Hall.
\
Hordijk, W., and Steel, M. (2018) Autocatalytic networks at the basis of life’s origin and organization. Life 8, no. 4, 62.
\
Erdős, P., and Rényi, A. (1960) On the evolution of random graphs. Publ. math.inst. hung. acad. sci 5, no. 1, 17-60.
\
Hordijk, W., and Steel, M. (2012) Autocatalytic sets extended: Dynamics, inhibition, and a generalization. Journal of Systems Chemistry 3, no. 1, 1-12.
\
Hordijk, W, Steel, M., Kauffman, S. A. (2012).The structure of autocatalytic sets: evolvability, enablement, and emergence. Acta Biotheoretica, Vol 60, Issue 4, 379-392.
\
Vassas. V., Fernando, C., Santos, M., Kauffman, S., Szathmary, E. (2012) Evolution Before Genes. Biology Direct. http://www.biology-direct.com/content/7/1/1
\
Puy, D. and Signore, M. (2002) From nuclei to atoms and molecules: the chemical history of the early universe. New Astronomy Reviews 46, no. 11, 709-723.
\
Kauffman, S. A., Jelenfi, D. Vattay, G. (2020) Theory of chemical evolution of molecule compositions in the universe, in the Miller-Urey experiment and the mass distribution of interstellar and intergalactic molecules J. Theor. Biol. Vol 486, https://doi.org/10.1016/j.jtbi.2019.110097.
\
Schmitt-Kopplin, P., Gabelica, Z., Gougeon, R., Hertkorn, N. (2010) High molecular diversity of extraterrestrial organic matter in Murchison meteorite revealed 40 years after its fall. Proceedings of the National Academy of Sciences 107, no. 7, 2763-276.
\
Steel,M., Hordijk W., Kauffman, S. A. (2020), Dynamics of a birth--death process based on combinatorial innovation, J. Theor. Biol 491, 11018.
\
Cortês, M., Kauffman, S. A., Liddle, A. R., Smolin L, Biocosmology and the theory of the adjacent possible. (2022) https://www.biocosmology.earth arXiv https://arxiv.org/abs/2204.14115
Koppl, R., Gatti, R., Deveraux, A., Fath, B., Herriot J., Hordijk, W., Kauffman S. A., Ulanowitcz, R., Valverde, S. (2023) Explaining Technology, Cambridge University Press, UK.
\
Hordijk, W. Koppl, R., Kauffman, S. A. (2023) Emergence of Autocatalytic Sets in a Simple Model of Technological Evolution, Journal of Evolutionary Economics, https://doi.org/10.1007/s00191-023-00838-2
Lane, N. (2015) Vital Question: Energy, Evolution, and the Origins of Complex Life. WW Norton & Company.
\
Damer, B., and Deamer, D. (2020) The hot spring hypothesis for an origin of life. Astrobiology 20, no. 4, 429-452.
\
Bergson, H. (1907). Creative Evolution. East India Publishing Company, Ottawa, Canada, 2022.
\
Wong, M., Cleland, C., Arend D., Bartlett, S., Cleaves, H., Demarest, H., Prabhu, A., Lunine, J. and Hazen R. (2023) On the roles of function and selection in evolving systems. Proceedings of the National Academy of Sciences 120, no. 43: e2310223120.
\
Smolin, L. (2013) Time reborn: From the crisis in physics to the future of the universe. HMH.
\
Kauffman, S. A. and Roli, A. (2021) The world is not a theorem. Entropy Vol 23, issue 11
\
Kauffman, S. A. and Roli, A. (2023) A third transition in science? Interface Focus13: 20220063. https://doi.org/10.1098/rsfs.2022.0063
\
Gould, S. J., and Vrba, E. S. (1982) Exaptation—a missing term in the science of form. Paleobiology 8, no. 1, 4-15.
\
Fraenkel, A., Bar-Hillel, Y. and Levy, A. (1973) Foundations of set theory. Elsevier.
\
Grattan-Guinness, I. (2000) The search for mathematical roots, 1870-1940: logics, set theories and the foundations of mathematics from Cantor through Russell to Gödel. Princeton University Press.
\
Skolem, Th. (1955) Peano's axioms and models of arithmetic. In Studies in Logic and the Foundations of Mathematics, vol. 16, pp. 1-14. Elsevier.
\
Cassidy, D. (1992) Heisenberg, uncertainty and the quantum revolution. Scientific American 266, no. 5, 106-113.
\
Exoplanet, Wikipedia
\
Krissansen-Totton, J., Thomson, M., Galloway, M., Fortney, J. (2022) Understanding planetary context to enable life detection on exoplanets and testing the Copernican Principle, Nature Astronomy, Vol 6, 189-198.
\
Wollrab, E., and Ott, A. (2018) A Miller–Urey broth mirrors the mass density distribution of all Beilstein indexed organic molecules. New Journal of Physics 20, no. 10 105003.
\
Lehman, N. E. and Kauffman, S. A, (2021), Constraint closure drove major transitions in the origins of life. Entropy, 23, 1, 105. https://doi.org/10.3390/e23010105
\
Carter C., Wills, P. (2017), Interdependence, Reflexivity, Fidelity, Impedence Matching, and the Evolution of Genetic Coding. Molecular Biology and Evolution, Vol 35, issue 2.
\
Kauffman, S. and Lehman, N. E. (2023) Mixed anhydrides at the intersection between peptide and RNA autocatalytic sets: evolution of biological coding, Journal of the Royal Society Interface. https://doi.org/10.1098/rsfs.2023.0009
\
Bordenave, G., Louis Pasteur (1822–1895). (2003) Microbes and infection 5, no. 6, 553-560.
\
Lazcano, A. (2016) Alexandr I. Oparin and the origin of life: a historical reassessment of the heterotrophic theory. Journal of molecular evolution 83, 214-222.
\
Tirard, S. (2017) JBS Haldane and the origin of life. Journal of genetics 96, no. 5, 735-739.
\
Miller, S. and Urey, H. (1959) Organic Compound Synthesis on the Primitive Earth. Science Vol 130 (3370) 245–51.
\
Macintyre, A. (2011) The impact of Gödel’s incompleteness theorems on mathematics. Kurt Gödel and the foundations of mathematics: Horizons of Truth 3-25.
\
\
:::info This paper is available on arxiv under CC BY 4.0 DEED license.
:::
\
This content originally appeared on HackerNoon and was authored by Homology
Homology | Sciencx (2024-08-18T21:00:34+00:00) Unlocking the Secrets of Autocatalytic Sets: How Bit Strings and Reactions Shape Molecular Evolution. Retrieved from https://www.scien.cx/2024/08/18/unlocking-the-secrets-of-autocatalytic-sets-how-bit-strings-and-reactions-shape-molecular-evolution/
Please log in to upload a file.
There are no updates yet.
Click the Upload button above to add an update.
